A Comprehensive Web Tool for Interpreting MS2 Data of Natural Products
About MassKG
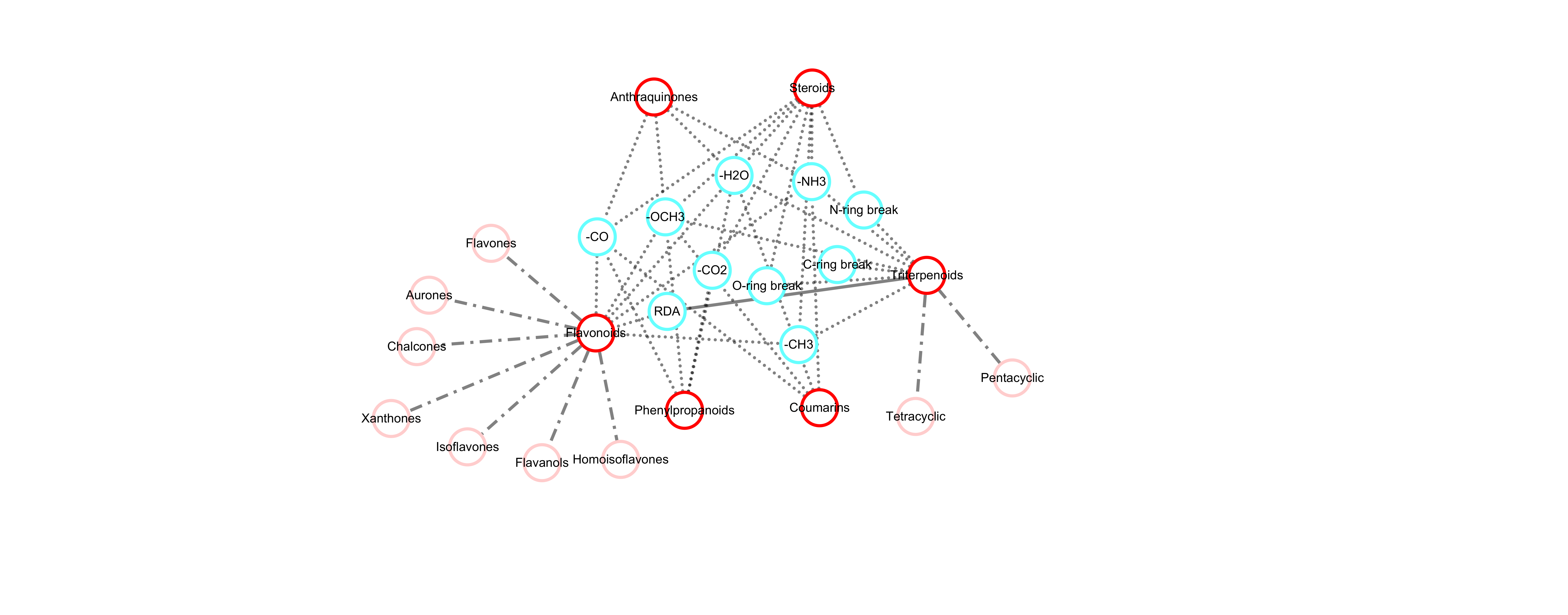
MassKG is designed to be an intelligent tool for the knowledge-based interpretation of metabolites based on MS data, with a particular focus on the fragmentation patterns of plant metabolites. We designed MassKG as a web server for a good availability of the aforementioned tools. In the current version, MassKG support both single spectra and batch MS file query modes as well as exploring the whole MassKG NP chemical space. It can be used in conjunction with MS data processing software like MZmine and MS-DIAL. After raw data processing, the yielded feature table containing precursor m/z and MS2 spectra can be used to as the inputs of MassKG web server.
Search
The single search tool consists of four blocks. First, the candidate database is to be selected. In this version, we have designed three databases: MassKG for the entire database, "coconut" for the known NPs, and "generated" for only the novel predicted NPs. The default database is MassKG. Next, the precursor mass and ion mode must be checked (area 1). The mass error tolerance block is set with a default value of 10 (mDa). The molecular formula block and adduct block (area 2) are not necessary since MassKG is able to calculate the candidate formula automatically. After that, users must input the MS2 table into the MS/MS text area (area 3). Once these procedures are finished, click the submit button (area4) and wait for a second. All the candidates will be gathered into a cleaned table, and their corresponding fragment trees will be constructed for direct visualization.
Here are examples for mgf file and msp file.

Batch
In order to streamline the handling of batch inference tasks,
we offer a comprehensive data reporting service. Users can upload MS data processed in formats like txt,msp files, or mgf.
Currently, the web service is testing so the user information is not required. You can skip these. We prepared two examples, here we use the example data to illustrate the batch analysis process.
1. Click the button and download the example mgf or msp file.Drag it to the upload area, the filename will be shown in that area.
2. Click the upload button, then the prompt of 'File uploaded sucessfully!' in green appears.
3. Select a ion mode。
4. Select a database, the default database is MassKG, which contains 400,000+ known NPs and 250,000+ novel structures.
5. Input a integer TopK value, which determines the final report will retain the top K candidates for each MS2 spectra. Default Topk is 5 and a value of 0 means return all candidates.
6. Input a threshold value, filtering peaks with area below the threshold. Default value is 0, means no peak filter.
7. Mass error tolerance set for inferring molecular formulas from precursor m/z values. Default is 0.005.
This module would return a metabolite annotation report.

REST API
MassKG now support a REST API for getting web service locally. The url is https://xomics.com.cn/masskg/Oneres, here we show a example of how to use this API through python.
import requests
import json
# set the input parameters
tolerence = "0.01"
mz = "783.4916"
mode = "-1"
formula = "C42H72O13"
ms2 = """780.98395 10
783.49719 1000
71.02712 7
59.02112 7
89.02495 7
179.05844 7
375.28809 7
459.38599 65
71.03307 5
73.03203 5
85.0288 5
131.03384 5
537.33856 5
119.03482 5
161.04524 54
783.35089 4
513.39459 4
555.40656 4
603.43585 3
619.44031 3"""
url = "https://xomics.com.cn/masskg/Oneres"
data = [
{"name": "miw", "value": mz},
{"name": "mode", "value": mode},
{"name": "ms2", "value": ms2},
{"name": "ppm", "value": tolerence},
{"name": "formula", "value": formula}
]
response = requests.post(url, data={'data': json.dumps(data)})
print(response.status_code)
print(response.json())
Running examples using python
All paramters should be str variable.
Tolerance The mass error tolerance, unit is mDa. A value between 0.001-0.01 is suggested.[default 0.005]
mz The precursor m/z. [required]
mode Ion mode, -1 is negative mode and 1 is positive mode. [required]
formula Molecule formula [optional]
ms2 MS2 table. fragment m/z and intensity should be devided by "\t" or space, and peaks should be seperated by "\n". We will update the support format of ms2 in near future.