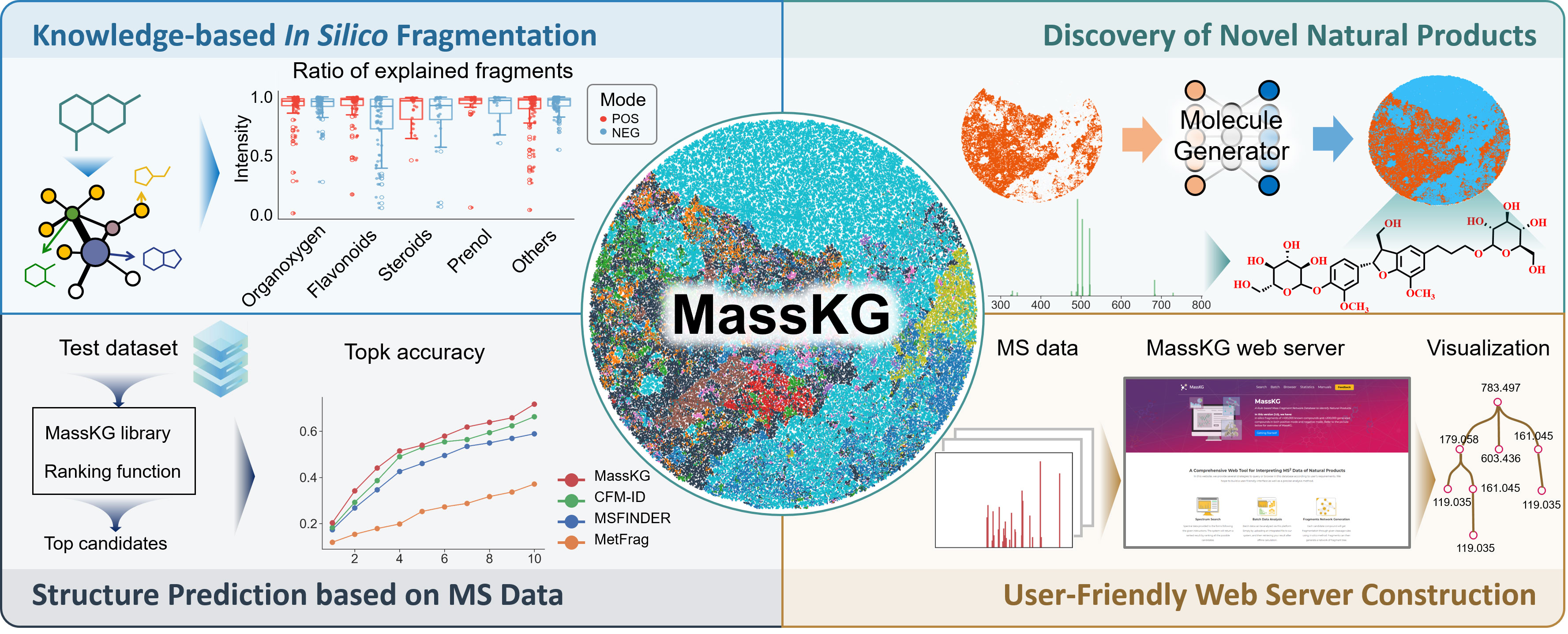
MassKG
MassKG
A Knowledge-based Mass Fragment Network Database to Identify Natural Products.
In this version (1.0), we have:
in-silico fragments of >400,000 known compounds and >200,000 generated compounds in
both positive mode and negative mode.
Refer to the picture below for overview of MassKG.
Our website will undergo maintenance and updates during Beijing time 10:00 PM to 2:00 AM every day.